Step 4 is contributed by Pierre Kestener. This step shows how to use AccFFT to solve a Poisson problem in parallel for both CPU and GPU. Pierre has also contributed an interface for writing/reading data using PNETCDF library.
Introduction
From molecular dynamics and quantum chemistry, to plasma physics and computational astrophysics, Poisson solvers are used in many applications in computational science and engineering. For a problem with periodic boundaries, Fourier methods are favorable due to their spectral accuracy and low computational cost to get such accuracy. Many of the interesting numerical problems involve billions of unknowns which certainly does not fit into shared memory. Therefore, one needs to compute distributed Fourier transforms. This is where AccFFT comes into the picture. This step shows you how to use AccFFT to solve the Poisson problem in parallel on both CPU and GPU. Moreover, you will see how Pierre’s utility function can be used to write the distributed data using PNETCDF library.
Poisson equation is defined as follows:
where can be thought of as the force distributed along a surface, and to be the resulting deformation. Now usually the force field is given and one needs to compute , which is given by:
For a simple geometry applying the inverse of the Laplace operator is straightforward with spectral analysis. If we take the Fourier transform of both sides we will have:
Here the hats refer to the variable in frequency domain, is the wave number, and is the inverse Fourier transform. In 3D the same thing applies and can be computed as follows:
Note that cannot be zero. This is because one cannot determine the constant value in without using additional data such as boundary conditions. This is evident since any satisfies the governing equation. For this reason we have to set in our numerical solver, and then calibrate the constant by using additional information.
So to recap:
- Compute FFT of
- Apply Poisson Fourier filter of
- Compute IFFT of the result
Code Explanation
The code is self explanatory and specially if you have read the previous steps you can easily understand what is going on by skimming thorough it. Here I want to comment on a new utility of function of AccFFT which you can use to write distributed data in a NetCDF file format for visualization. For example in the code you will find the following call to write the numerical result for :
1
2
3
4
std::string filename = "phi.nc";
MPI_Offset istart_mpi[3] = { istart[0], istart[1], istart[2] };
MPI_Offset isize_mpi[3] = { isize[0], isize[1], isize[2] };
write_pnetcdf(filename,istart_mpi,isize_mpi,n, phi);
You need to pass the output file name, the istart offset and the size of the data that resides in each process (computed for you by calling accfft_local_size_dft_* function), the global sizes, and of course a pointer to the data itself. The output will be a NetCDF file that can be read by a myriad of visualization softwares including Paraview.
Results
There are three test cases in the code for the force function :
- Test case 0:
- Test case 1:
- Test case 2:
Test case 0, is a harmonic force, test case 1 is an exponential force which becomes sharper as increases, and test case 2 is a discontinuous spherical force. You can specify the test case by passing “-t #” to the executable. The complete options are as follows:
- x,y,z: global domain sizes.
- t: testcase
- m: method
- a,b,c,r: uniform ball parameter (center location + radius)
- l:
Now the method option, m, is by default the method explained above. However, you can use a different approach (still using Fourier method). Instead of taking a Fourier transform from both sides of the Laplace operator, one can use Finite Differences to compute derivatives and then compute the FFT. Then from the orthogonality of the Fourier basis you can compute the Fourier coefficients of . This method has a lower accuracy but it is nice to see how it works. For details please see section 19.1 and 19.4 of the numerical bible.
Alright, so on Maverick I get the following results:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
ibrun -np 4 ./step4 -x 128 -y 128 -z 128 -m 1
---> Using test case number : 0
---> Using Fourier filter method 1
ERROR c_dims!=nprocs --> 0*0 !=4
Switching to c_dims_0=2 , c_dims_1=2
[mpi rank 1] isize 64 64 128 osize 128 64 32
[mpi rank 2] isize 64 64 128 osize 128 64 33
[mpi rank 1] istart 0 64 0 ostart 0 0 33
[mpi rank 2] istart 64 0 0 ostart 0 64 0
[mpi rank 0] isize 64 64 128 osize 128 64 33
[mpi rank 3] isize 64 64 128 osize 128 64 32
[mpi rank 0] istart 0 0 0 ostart 0 0 0
[mpi rank 3] istart 64 64 0 ostart 0 64 33
#################################################################
L2 relative error between phi and exact solution : 1.28362e-31
#################################################################
Timing for FFT of size 128*128*128
Setup 0.543655
FFT 0.0114319
IFFT 0.00930095
And with the Finite Difference method:
1
2
3
4
5
6
7
8
9
10
11
ibrun -np 4 ./step4 -x 128 -y 128 -z 128 -m 0
.
.
.
#################################################################
L2 relative error between phi and exact solution : 4.03294e-08
#################################################################
Timing for FFT of size 128*128*128
Setup 0.536285
FFT 0.0117891
IFFT 0.00952101
As you can see the full spectral method gets a better accuracy in this case. I suggest you try other test cases and for example vary the radius of the discontinuous spherical force in test case 2, or vary in test case 1 and compare the results.
If you have installed AccFFT with PNETCDF you should see three netCDF files in the step4 directory. These include the force term written to rho.nc, the exact phi written to phi_exact.nc, and the numerically computed phi written to phi.nc. If you have Paraview installed on your machine, you can load these directly by using “NetCDF files generic and CF conventions” plug-in.



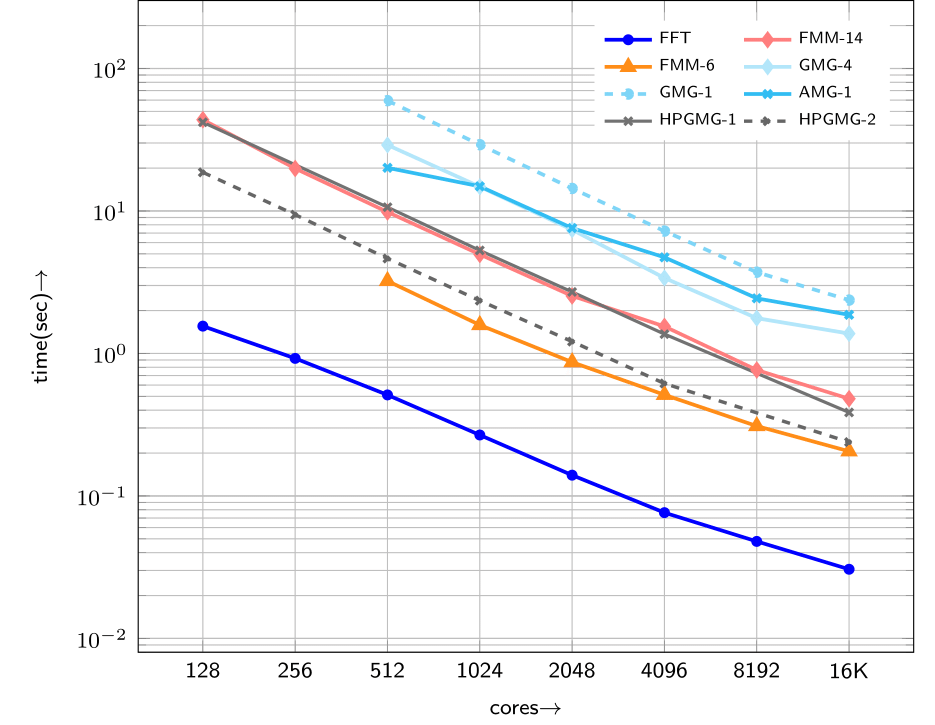
Comparison with FEM and FMM
In case you were interested in how this problem can be solved with Finited Element Method or Fast Multipole Method, be sure to check out this paper. We have compared parallel performance and accuracy of these methods with the spectral one. Test case 0 and a variant of test case 1 are used with different exponential powers. The conclusion of the study is that for a target accuracy, the spectral method is faster with a large margin compared to FEM/FMM. This is true, unless the force term is extremely sharp. In that case, FEM/FMM may be faster because they can use adaptive discretization, while the classical FFT can only use uniform mesh.
Complete CPU Code
Here is the Poisson solver of the CPU code. The full code is available here.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
801
802
803
804
805
806
807
808
809
810
811
812
813
814
815
/*
* File: step4.cpp
* Project: AccFFT
* Created by Amir Gholami on 12/23/2014
* Contact: contact@accfft.org
* Copyright (c) 2014-2015
*
* A very simple test for solving Laplacian(\phi) = rho using FFT in 3D.
*
* Laplacian operator can be considered as a low-pass filter.
* Here we implement 2 types of filters :
*
* method 0 : see Numerical recipes in C, section 19.4
* method 1 : just divide right hand side by -(kx^2+ky^2+kz^2) in Fourier
*
* Test case 0: rho(x,y,z) = sin(2*pi*x/Lx)*sin(2*pi*y/Ly)*sin(2*pi*z/Lz)
* Test case 1: rho(x,y,z) = (4*alpha*alpha*(x^2+y^2+z^2)-6*alpha)*exp(-alpha*(x^2+y^2+z^2))
* Test case 2: rho(x,y,z) = ( r=sqrt(x^2+y^2+z^2) < R ) ? 1 : 0
*
* Example of use:
* ./step4 -x 64 -y 64 -z 64 -m 1 -t 2
*
* \author Pierre Kestener
* \date June 1st, 2015
*/
#include <mpi.h>
#include <stdlib.h>
#include <math.h> // for M_PI
#include <unistd.h> // for getopt
#include <accfft.h>
#include <accfft_utils.h>
#include <string>
#define SQR(x) ((x)*(x))
enum TESTCASE {
TESTCASE_SINE=0,
TESTCASE_GAUSSIAN=1,
TESTCASE_UNIFORM_BALL=2
};
struct PoissonParams {
// global domain sizes
int nx;
int ny;
int nz;
// there 3 testcases
int testcase;
// method number: 2 variants (see head of this file)
int methodNb;
// only valid for the uniform ball testcase (coordinate of center of the ball)
double xC;
double yC;
double zC;
double radius;
// only valid for the gaussian testcase
double alpha;
// define constructor for default values
PoissonParams() :
nx(128),
ny(128),
nz(128),
testcase(TESTCASE_SINE),
methodNb(0),
xC(0.5),
yC(0.5),
zC(0.5),
radius(0.1),
alpha(30.0)
{
} // PoissonParams::PoissonParams
}; // struct PoissonParams
// =======================================================
// =======================================================
/*
* RHS of testcase sine: eigenfunctions of Laplacian
*/
double testcase_sine_rhs(double x, double y, double z) {
return sin(2*M_PI*x) * sin(2*M_PI*y) * sin(2*M_PI*z);
} // testcase_sine_rhs
// =======================================================
// =======================================================
/*
* Solution of testcase sine: eigenfunctions of Laplacian
*/
double testcase_sine_sol(double x, double y, double z) {
return -sin(2*M_PI*x) * sin(2*M_PI*y) * sin(2*M_PI*z) / ( 3*(4*M_PI*M_PI) );
} // testcase_sine_sol
// =======================================================
// =======================================================
/*
* RHS of testcase gaussian
*/
double testcase_gaussian_rhs(double x, double y, double z, double alpha) {
return (4*alpha*alpha*(x*x+y*y+z*z)-6*alpha)*exp(-alpha*(x*x+y*y+z*z));
} // testcase_gaussian_rhs
// =======================================================
// =======================================================
/*
* Solution of testcase gaussian
*/
double testcase_gaussian_sol(double x, double y, double z, double alpha) {
return exp(-alpha*(x*x+y*y+z*z));
} // testcase_gaussian_sol
// =======================================================
// =======================================================
/*
* RHS of testcase uniform ball
*/
double testcase_uniform_ball_rhs(double x, double y, double z,
double xC, double yC, double zC,
double R) {
double r = sqrt( (x-xC)*(x-xC) + (y-yC)*(y-yC) + (z-zC)*(z-zC) );
double res = r < R ? 1.0 : 0.0;
return res;
} // testcase_uniform_ball_rhs
// =======================================================
// =======================================================
/*
* Solution of testcase uniform ball
*/
double testcase_uniform_ball_sol(double x, double y, double z,
double xC, double yC, double zC,
double R) {
double r = sqrt( (x-xC)*(x-xC) + (y-yC)*(y-yC) + (z-zC)*(z-zC) );
double res = r < R ? r*r/6.0 : -R*R*R/(3*r)+R*R/2;
return res;
} // testcase_uniform_ball_sol
// =======================================================
// =======================================================
/*
* Initialize the rhs of Poisson equation, and exact known solution.
*
* \param[out] rho Poisson rhs array
* \param[out] sol known exact solution to corresponding Poisson problem
*/
template<const TESTCASE testcase_id>
void initialize(double *rho, double *sol, int *n, MPI_Comm c_comm, PoissonParams ¶ms)
{
double pi=M_PI;
int n_tuples=n[2];
int istart[3], isize[3], osize[3],ostart[3];
accfft_local_size_dft_r2c(n,isize,istart,osize,ostart,c_comm);
/*
* testcase gaussian parameters
*/
double alpha=1.0;
if (testcase_id == TESTCASE_GAUSSIAN)
alpha = params.alpha;
/*
* testcase uniform ball parameters
*/
// uniform ball function center
double xC = params.xC;
double yC = params.yC;
double zC = params.zC;
// uniform ball radius
double R = params.radius;
{
double X,Y,Z;
double x0 = 0.5;
double y0 = 0.5;
double z0 = 0.5;
long int ptr;
for (int i=0; i<isize[0]; i++) {
for (int j=0; j<isize[1]; j++) {
for (int k=0; k<isize[2]; k++) {
X = 1.0*(i+istart[0])/n[0];
Y = 1.0*(j+istart[1])/n[1];
Z = 1.0*(k+istart[2])/n[2];
ptr = i*isize[1]*isize[2] + j*isize[2] + k;
if (testcase_id == TESTCASE_SINE) {
rho[ptr] = testcase_sine_rhs(X,Y,Z);
sol[ptr] = testcase_sine_sol(X,Y,Z);
} else if (testcase_id == TESTCASE_GAUSSIAN) {
rho[ptr] = testcase_gaussian_rhs(X-x0,Y-x0,Z-x0,alpha);
sol[ptr] = testcase_gaussian_sol(X-x0,Y-x0,Z-x0,alpha);
} else if (testcase_id == TESTCASE_UNIFORM_BALL) {
rho[ptr] = testcase_uniform_ball_rhs(X,Y,Z,xC,yC,zC,R);
sol[ptr] = testcase_uniform_ball_sol(X,Y,Z,xC,yC,zC,R);
}
} // end for k
} // end for j
} // end for i
/*
* rescale exact solution to ease comparison with numerical solution which has a zero average value
*/
if (testcase_id == TESTCASE_UNIFORM_BALL) {
// make exact solution allways positive to ease comparison with numerical one
// compute local min
double minVal = sol[0];
for (int i=0; i<isize[0]; i++){
for (int j=0; j<isize[1]; j++){
for (int k=0; k<isize[2]; k++){
ptr = i*isize[1]*n_tuples + j*n_tuples + k;
if (sol[ptr] < minVal)
minVal = sol[ptr];
} // end for k
} // end for j
} // end for i
// compute global min
double minValG;
MPI_Allreduce(&minVal, &minValG, 1, MPI_DOUBLE, MPI_MIN, MPI_COMM_WORLD);
for (int i=0; i<isize[0]; i++){
for (int j=0; j<isize[1]; j++){
for (int k=0; k<isize[2]; k++){
ptr = i*isize[1]*n_tuples + j*n_tuples + k;
sol[ptr] -= minValG;
} // end for k
} // end for j
} // end for i
} // end TESTCASE_UNIFORM_BALL
}
return;
} // end initialize
// =======================================================
// =======================================================
/*
* Poisson fourier filter.
* Divide fourier coefficients by -(kx^2+ky^2+kz^2).
*/
void poisson_fourier_filter(Complex *data_hat,
int N[3],
int isize[3],
int istart[3],
int methodNb) {
int nprocs, procid;
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
MPI_Comm_size(MPI_COMM_WORLD, &nprocs);
double NX = N[0];
double NY = N[1];
double NZ = N[2];
double Lx = 1.0;
double Ly = 1.0;
double Lz = 1.0;
double dx = Lx/NX;
double dy = Ly/NY;
double dz = Lz/NZ;
for (int i=0; i < isize[0]; i++) {
for (int j=0; j < isize[1]; j++) {
for (int k=0; k < isize[2]; k++) {
double kx = istart[0]+i;
double ky = istart[1]+j;
double kz = istart[2]+k;
double kkx = (double) kx;
double kky = (double) ky;
double kkz = (double) kz;
if (kx>NX/2)
kkx -= NX;
if (ky>NY/2)
kky -= NY;
if (kz>NZ/2)
kkz -= NZ;
int index = i*isize[1]*isize[2]+j*isize[2]+k;
double scaleFactor = 0.0;
if (methodNb==0) {
/*
* method 0 (See Eq. 19.4.5 of Numerical recipes 2nd Ed.)
*/
scaleFactor=2*(
(cos(1.0*2*M_PI*kx/NX) - 1)/(dx*dx) +
(cos(1.0*2*M_PI*ky/NY) - 1)/(dy*dy) +
(cos(1.0*2*M_PI*kz/NZ) - 1)/(dz*dz) );
} else if (methodNb==1) {
/*
* method 1 (just from Continuous Fourier transform of
* Poisson equation)
*/
//scaleFactor=-4*M_PI*M_PI*(kkx*kkx + kky*kky + kkz*kkz)/;
scaleFactor=-(4*M_PI*M_PI/Lx/Lx*kkx*kkx + 4*M_PI*M_PI/Ly/Ly*kky*kky + 4*M_PI*M_PI/Ly/Ly*kkz*kkz);
}
scaleFactor*=NX*NY*NZ; // FFT scaling factor
if (kx!=0 or ky!=0 or kz!=0) {
data_hat[index][0] /= scaleFactor;
data_hat[index][1] /= scaleFactor;
} else { // enforce mean value is zero since you cannot recover the zero frequency
data_hat[index][0] = 0.0;
data_hat[index][1] = 0.0;
}
}
}
}
} // poisson_fourier_filter
// =======================================================
// =======================================================
/*
* Rescale Numerical Solution.
* The zeroth frequency data (i.e. the mean of the solution)
* cannot be recovered in the Poisson problem without using additional
* data, such as boundary condition. Without such information
* any u=u+constant would satisfy the problem.
*
* Here we use specific information for each test case
* to "calibrate" the numerical solution with the
* correct constant.
*/
void rescale_numerical_solution(double *phi,
int *isize,
PoissonParams ¶ms) {
const int testcase_id = params.testcase;
int n_tuples=params.nz;
if (testcase_id == TESTCASE_GAUSSIAN) {
// compute local max
double maxVal = phi[0];
for (int index=0;
index < isize[0]*isize[1]*isize[2];
index++) {
if (phi[index] > maxVal)
maxVal = phi[index];
} // end for index
// compute global max
double maxValG;
MPI_Allreduce(&maxVal, &maxValG, 1, MPI_DOUBLE, MPI_MAX, MPI_COMM_WORLD);
for (int index=0;
index < isize[0]*isize[1]*isize[2];
index++) {
phi[index] += 1-maxValG;
} // end for index
} // end TESTCASE_GAUSSIAN
if (testcase_id == TESTCASE_UNIFORM_BALL) {
// make exact solution allways positive to ease comparison with numerical one
// compute local min
double minVal = phi[0];
for (int index=0;
index < isize[0]*isize[1]*isize[2];
index++) {
if (phi[index] < minVal)
minVal = phi[index];
}
// compute global min
double minValG;
MPI_Allreduce(&minVal, &minValG, 1, MPI_DOUBLE, MPI_MIN, MPI_COMM_WORLD);
for (int index=0;
index < isize[0]*isize[1]*isize[2];
index++) {
phi[index] -= minValG;
}
} // end TESTCASE_UNIFORM_BALL
} // rescale_numerical_solution
// =======================================================
// =======================================================
void compute_L2_error(double *phi,
double *phi_exact,
int *isize,
PoissonParams ¶ms) {
int nprocs, procid;
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
MPI_Comm_size(MPI_COMM_WORLD, &nprocs);
/* global domain size */
int n[3];
n[0] = params.nx;
n[1] = params.ny;
n[2] = params.nz;
int n_tuples=n[2];
int N=n[0]*n[1]*n[2];
// compute L2 difference between FFT-based solution (phi) and
// expected analytical solution
double L2_diff = 0.0;
double L2_phi = 0.0;
// global values
double L2_diff_G = 0.0;
double L2_phi_G = 0.0;
long int ptr;
for (int index=0;
index < isize[0]*isize[1]*isize[2];
index++) {
L2_phi += phi_exact[index]*phi_exact[index];
L2_diff += (phi[index]-phi_exact[index])*(phi[index]-phi_exact[index]);
}
// global L2
MPI_Reduce(&L2_phi, &L2_phi_G, 1, MPI_DOUBLE, MPI_SUM, 0, MPI_COMM_WORLD);
MPI_Reduce(&L2_diff, &L2_diff_G, 1, MPI_DOUBLE, MPI_SUM, 0, MPI_COMM_WORLD);
if (procid==0) {
std::cout << "#################################################################" << std::endl;
std::cout << "L2 relative error between phi and exact solution : "
<< L2_diff_G / L2_phi_G
//<< " ( = " << L2_diff_G << "/" << L2_phi_G << ")"
<< std::endl;
std::cout << "#################################################################" << std::endl;
}
} // compute_L2_error
// =======================================================
// =======================================================
/*
* FFT-based poisson solver.
*
* \param[in] params parameters parsed from the command line arguments
* \param[in] nthreads number of threads
*/
void poisson_solve(PoissonParams ¶ms, int nthreads) {
int nprocs, procid;
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
MPI_Comm_size(MPI_COMM_WORLD, &nprocs);
// which testcase
const int testCaseNb = params.testcase;
// which method ? variant of FFT-based Poisson solver : 0 or 1
const int methodNb = params.methodNb;
if (procid==0)
printf("---> Using Fourier filter method %d\n",methodNb);
/* global domain size */
int n[3];
n[0] = params.nx;
n[1] = params.ny;
n[2] = params.nz;
/* Create Cartesian Communicator */
int c_dims[2] = {0};
MPI_Comm c_comm;
accfft_create_comm(MPI_COMM_WORLD,c_dims,&c_comm);
//printf("[mpi rank %d] c_dims = %d %d\n", procid, c_dims[0], c_dims[1]);
// Governing Equation: Laplace(\phi)=rho
double *rho, *exact_solution;
Complex *phi_hat;
double f_time=0*MPI_Wtime(),i_time=0, setup_time=0;
int alloc_max=0;
int isize[3],osize[3],istart[3],ostart[3];
/* Get the local pencil size and the allocation size */
alloc_max=accfft_local_size_dft_r2c(n,isize,istart,osize,ostart,c_comm);
printf("[mpi rank %d] isize %3d %3d %3d osize %3d %3d %3d\n", procid,
isize[0],isize[1],isize[2],
osize[0],osize[1],osize[2]
);
printf("[mpi rank %d] istart %3d %3d %3d ostart %3d %3d %3d\n", procid,
istart[0],istart[1],istart[2],
ostart[0],ostart[1],ostart[2]
);
rho=(double*)accfft_alloc(isize[0]*isize[1]*isize[2]*sizeof(double));
phi_hat=(Complex*)accfft_alloc(alloc_max);
exact_solution=(double*)accfft_alloc(isize[0]*isize[1]*isize[2]*sizeof(double));
accfft_init(nthreads);
setup_time=-MPI_Wtime();
/* Create FFT plan */
accfft_plan * plan = accfft_plan_dft_3d_r2c(n,
rho, (double*)phi_hat,
c_comm, ACCFFT_MEASURE);
setup_time+=MPI_Wtime();
/* Initialize rho (force) */
switch(testCaseNb) {
case TESTCASE_SINE:
initialize<TESTCASE_SINE>(rho, exact_solution, n, c_comm, params);
break;
case TESTCASE_GAUSSIAN:
initialize<TESTCASE_GAUSSIAN>(rho, exact_solution, n, c_comm, params);
break;
case TESTCASE_UNIFORM_BALL:
initialize<TESTCASE_UNIFORM_BALL>(rho, exact_solution, n, c_comm, params);
break;
}
MPI_Barrier(c_comm);
// optional : save rho (rhs)
#ifdef USE_PNETCDF
{
std::string filename = "rho.nc";
MPI_Offset istart_mpi[3] = { istart[0], istart[1], istart[2] };
MPI_Offset isize_mpi[3] = { isize[0], isize[1], isize[2] };
write_pnetcdf(filename,
istart_mpi,
isize_mpi,
c_comm,
n,
rho);
}
#else
{
if (procid==0)
std::cout << "[WARNING] You have to enable PNETCDF to be enable to dump data into files\n";
}
#endif // USE_PNETCDF
/*
* Perform forward FFT
*/
f_time-=MPI_Wtime();
accfft_execute_r2c(plan,rho,phi_hat);
f_time+=MPI_Wtime();
MPI_Barrier(c_comm);
/*
* here perform fourier filter associated to poisson ...
*/
poisson_fourier_filter(phi_hat, n, osize, ostart, methodNb);
/*
* Perform backward FFT
*/
double * phi=(double*)accfft_alloc(isize[0]*isize[1]*isize[2]*sizeof(double));
i_time-=MPI_Wtime();
accfft_execute_c2r(plan,phi_hat,phi);
i_time+=MPI_Wtime();
/* rescale numerical solution before computing L2 */
rescale_numerical_solution(phi, isize, params);
/* L2 error between phi and phi_exact */
compute_L2_error(phi, exact_solution, isize, params);
/* optional : save phi (solution to poisson) and exact solution */
#ifdef USE_PNETCDF
{
std::string filename = "phi.nc";
MPI_Offset istart_mpi[3] = { istart[0], istart[1], istart[2] };
MPI_Offset isize_mpi[3] = { isize[0], isize[1], isize[2] };
write_pnetcdf(filename,
istart_mpi,
isize_mpi,
c_comm,
n,
phi);
}
{
std::string filename = "phi_exact.nc";
MPI_Offset istart_mpi[3] = { istart[0], istart[1], istart[2] };
MPI_Offset isize_mpi[3] = { isize[0], isize[1], isize[2] };
write_pnetcdf(filename,
istart_mpi,
isize_mpi,
c_comm,
n,
exact_solution);
}
#else
{
if (procid==0)
std::cout << "[WARNING] You have to enable PNETCDF to be enable to dump data into files\n";
}
#endif // USE_PNETCDF
/* Compute some timings statistics */
double g_f_time, g_i_time, g_setup_time;
MPI_Reduce(&f_time,&g_f_time,1, MPI_DOUBLE, MPI_MAX,0, MPI_COMM_WORLD);
MPI_Reduce(&i_time,&g_i_time,1, MPI_DOUBLE, MPI_MAX,0, MPI_COMM_WORLD);
MPI_Reduce(&setup_time,&g_setup_time,1, MPI_DOUBLE, MPI_MAX,0, MPI_COMM_WORLD);
PCOUT<<"Timing for FFT of size "<<n[0]<<"*"<<n[1]<<"*"<<n[2]<<std::endl;
PCOUT<<"Setup \t"<<g_setup_time<<std::endl;
PCOUT<<"FFT \t"<<g_f_time<<std::endl;
PCOUT<<"IFFT \t"<<g_i_time<<std::endl;
accfft_free(rho);
accfft_free(exact_solution);
accfft_free(phi_hat);
accfft_free(phi);
accfft_destroy_plan(plan);
accfft_cleanup();
MPI_Comm_free(&c_comm);
return;
} // end poisson_solve
// =======================================================
// =======================================================
/*
* Read poisson parameters from command line argument.
*
* \param[in] argc
* \param[in] argv
* \param[out] params a reference to a PoissonParams structure.
*
* options:
* - x,y,z are for global domain sizes.
* - t for testcase
* - m for method
* - a,b,c,r for uniform ball parameter (center location + radius)
* - l for alpha
*/
void getPoissonParams(const int argc, char *argv[],
PoissonParams ¶ms) {
int nprocs, procid;
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
MPI_Comm_size(MPI_COMM_WORLD, &nprocs);
//opterr = 0;
char *value = NULL;
int c;
int tmp;
/*
*
*/
while ((c = getopt (argc, argv, "x:y:z:m:t:a:b:c:r:l:")) != -1)
switch (c)
{
case 'x':
value = optarg;
params.nx = atoi(value);
break;
case 'y':
value = optarg;
params.ny = atoi(value);
break;
case 'z':
value = optarg;
params.nz = atoi(value);
break;
case 'm':
value = optarg;
tmp = atoi(value);
if (tmp < 0 || tmp > 1) {
// wrong value, defaulting to 1
tmp = 1;
if (procid==0) std::cout << "wrong value for option -m (method); defaulting to 0\n";
}
params.methodNb = tmp;
break;
case 't':
value = optarg;
tmp = atoi(value);
if (tmp < 0 || tmp > 2) {
// wrong value, defaulting to 0
tmp = 0;
if (procid==0) std::cout << "wrong value for option -t (testcase); defaulting to 0\n";
}
params.testcase = tmp;
break;
case 'a':
value = optarg;
params.xC = atof(value);
break;
case 'b':
value = optarg;
params.yC = atof(value);
break;
case 'c':
value = optarg;
params.zC = atof(value);
break;
case 'r':
value = optarg;
params.radius = atof(value);
break;
case 'l':
value = optarg;
params.alpha = atof(value);
break;
case '?':
if (procid==0) std::cerr << "#### All options require an argument. ####\n";
default:
;
}
} // getPoissonParams
/******************************************************/
/******************************************************/
/******************************************************/
int main(int argc, char *argv[])
{
MPI_Init (&argc, &argv);
int nprocs, procid;
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
MPI_Comm_size(MPI_COMM_WORLD, &nprocs);
/* parse command line arguments and fill params structure */
PoissonParams params = PoissonParams();
getPoissonParams(argc, argv, params);
// test case number
const int testCaseNb = params.testcase;
if (testCaseNb < 0 || testCaseNb > 2) {
if (procid == 0) {
std::cerr << "---> Wrong test case. Must be integer < 2 !!!\n";
}
} else {
if (procid == 0) {
std::cout << "---> Using test case number : " << testCaseNb << std::endl;
}
}
int nthreads=1;
poisson_solve(params, nthreads);
MPI_Finalize();
return 0;
} // end main